E. Coli pangenome analysis¶
To test our tool, we decided to annotate Escherichia Coli pangenome. Escherichia Coli is a well-studied
bacterial species. It is a facultative anaerobic, Gram-negative, rod-shaped bacterium abundant in human gut
microbiome. It colonizes the gut within hours after birth and coexist with human throughout whole life1.
It also may exhibit pathogenic properties if it acquires virulence factors.
We used a pipeline that consists of kneaddata,
MegaHIT2, MetaBAT23, CheckM4,
and GTDB-tk5, for assembly, quality control, taxonomical annotation of
metagenome-assembled genomes (MAGs). We used Prodigal6 to predict genes and clustered
sequences by 95% similarity and 90% alignment coverage using CD-HIT7 to generate
non-redundant gene catalogue. This procedure yielded 11,813 protein sequences.
Metagenomic-DeepFRI uses 2 inputs: protein sequence and protein structure. As we obtained the protein sequences
in the previous step, we still do not have the protein structure. Luckily, after running update_target_mmseqs_database.py
the pipeline takes care of it. It searches for the similar sequence in supplied database using Bit-score. If similar sequences
exists in database, it sends the structure as the input to a graph-convolutional neural network (GCN). If there is no such sequence,
the prediction is made with help of convolutional neural network (CNN).
We tested the influence of database on the DeepFRI predictions. We used the following databases:
Rosetta
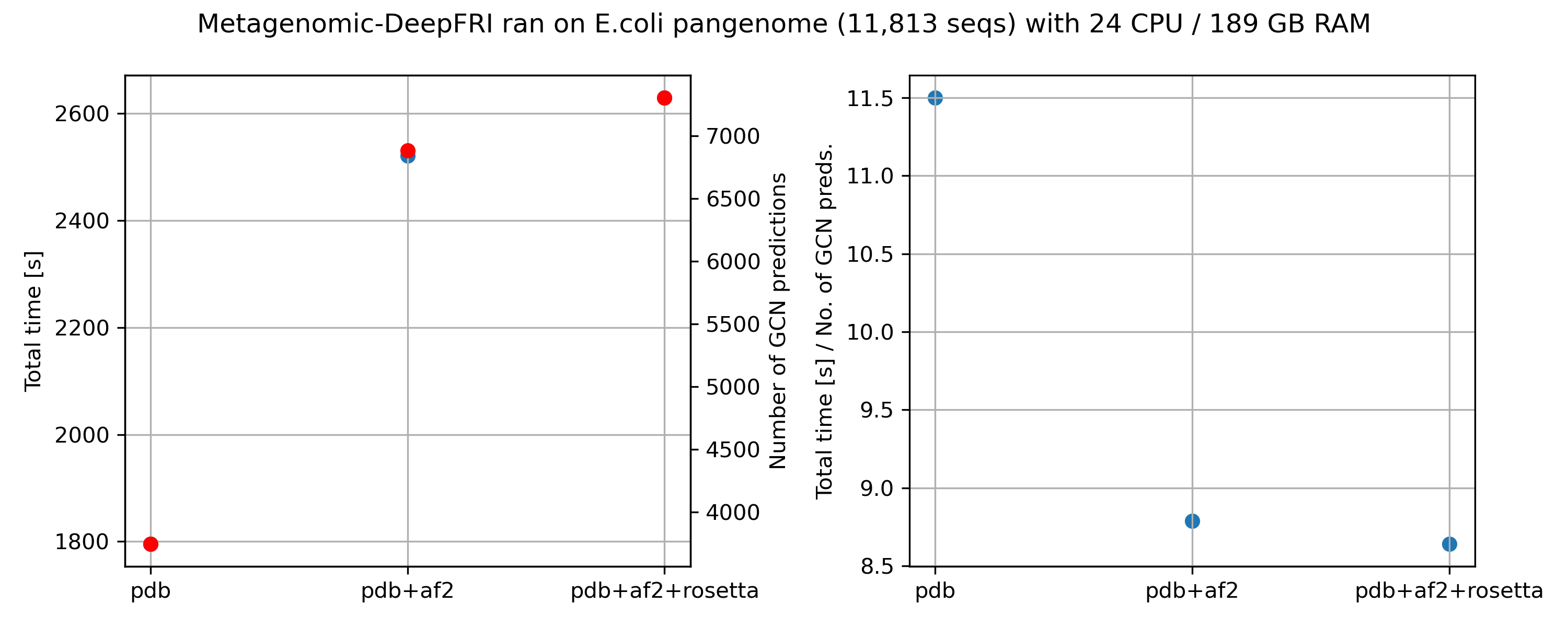
Next, we took a closer look on predicted GO terms. We excluded predictions with confidence < 0.2. We compared predictions with and without structural input.
We defined informative GO terms that satisfy the following parameters:
Terms that are associated with more than k=2,000 proteins
Rach of their descendant term contains less than 2,000 proteins (k=2,000 equates to approximately 1 of every 5,000 UniRef50 protein families).
Method |
No structural input |
PDB + AF2 + Rosetta |
|---|---|---|
No. unfiltered GO terms |
11,726 (99%) |
7,219 (61%) |
Informative GO terms |
2,016 (17%) |
3,012 (25%) |
The number of total predicted GO terms decreased, but the number of informative GO terms increased.
References
- 1
James B. Kaper, James P. Nataro, and Harry L. T. Mobley. Pathogenic escherichia coli. Nature Reviews Microbiology, 2(2):123–140, Feb 2004. URL: https://doi.org/10.1038/nrmicro818, doi:10.1038/nrmicro818.
- 2
Dinghua Li, Chi-Man Liu, Ruibang Luo, Kunihiko Sadakane, and Tak-Wah Lam. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics, 31(10):1674–1676, 01 2015. URL: https://doi.org/10.1093/bioinformatics/btv033, arXiv:https://academic.oup.com/bioinformatics/article-pdf/31/10/1674/17085710/btv033.pdf, doi:10.1093/bioinformatics/btv033.
- 3
Dongwan Kang, Feng Li, Edward S Kirton, Ashleigh Thomas, Rob S Egan, Hong An, and Zhong Wang. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ, February 2019.
- 4
Donovan H Parks, Michael Imelfort, Connor T Skennerton, Philip Hugenholtz, and Gene W Tyson. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res., 25(7):1043–1055, July 2015.
- 5
Pierre-Alain Chaumeil, Aaron J Mussig, Philip Hugenholtz, and Donovan H Parks. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics, November 2019.
- 6
Doug Hyatt, Gwo-Liang Chen, Philip F Locascio, Miriam L Land, Frank W Larimer, and Loren J Hauser. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics, 11(1):119, March 2010.
- 7
Limin Fu, Beifang Niu, Zhengwei Zhu, Sitao Wu, and Weizhong Li. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics, 28(23):3150–3152, December 2012.